Welcome to the Epromoter Website
Go to Clustering ToolEpromoters
Definition
Epromoters are defined as coding genes promoters that harbor both promoter and enhancer activity.
Background
Gene transcription in higher eukaryotes relies on a diverse network of regulatory elements, including promoters and enhancers, which traditionally have been seen as distinct types of regulatory elements. Promoters, located near the transcription start site (TSS), initiate local gene transcription, whereas enhancers, positioned farther from the TSS, activate gene expression over larger distances.
However, their distinction has become more blurred in recent years notably by the discovery of Epromoters, cis-regulatory sequences that combine architectural and functional characteristics of both promoters and enhancers. Epromoters can regulate distal promoters when assessed in episomal reporter systems or high-throughput reporter assays in different cellular contexts from Drosophila to humans, while their deletion or epigenetic silencing in their natural context results in the loss of expression of distal genes. Moreover, genetic variation within Epromoters can potentially influence the expression of distal genes.
Key Highlights

Resources
We have compiled a comprehensive resource of Epromoters based on STARR-seq experiments in multiple human cell lines and conditions.
Link to the Epromoters dataset: Download Supplemental Table 3
This file contains the directly identified Epromoter data from our study. For additional data and supplementary information, please refer to the publication's supplementary files.
*If you use this resource for a publication, please cite the following reference: Wan et al. Nucleic Acids Res. Dec 27:gkae1270
Key Publications
- Zabidi, M.A. et al. (2015) Enhancer–core-promoter specificity separates developmental and housekeeping gene regulation. Nature, 518, 556-559.
- Nguyen, T.A. et al. (2016) High-throughput functional comparison of promoter and enhancer activities. Genome Research, 26, 1023-1033.
- Engreitz, J.M. et al. (2016) Local regulation of gene expression by lncRNA promoters, transcription, and splicing. Nature, 539, 452-455.
- Dao, L.T.M. et al. (2017) Genome-wide characterization of mammalian promoters with distal enhancer functions. Nat Genet, 49, 1073-1081.
- Diao, Y. et al. (2017) A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nature Methods, 14, 629-635.
- Santiago-Algarra, D. et al. (2021) Epromoters function as a hub to recruit key transcription factors required for the inflammatory response. Nature Communications, 12, 6660.
- Malfait, J., Wan, J. and Spicuglia, S. (2023) Epromoters are new players in the regulatory landscape with potential pleiotropic roles. Bioessays, 45, e2300012.
- Mitchelmore, J. et al. (2020) Functional effects of variation in transcription factor binding highlight long-range gene regulation by Epromoters. Nucleic Acids Research, 48, 2866-2879.
- Jung, I. et al. (2019) A compendium of promoter-centered long-range chromatin interactions in the human genome. Nat Genet, 51, 1442-1449.
- Wan, J. et al. (2024) Comprehensive mapping of genetic variation at Epromoters reveals pleiotropic association with multiple disease traits. Nucleic Acids Res. Dec 27:gkae1270.
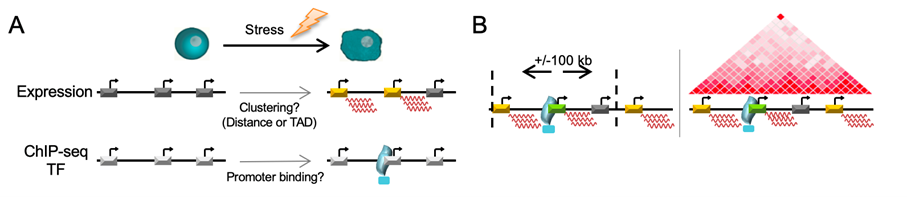
Clustering Tool
We have developed a pipeline to infer clusters of stress-induced genes potentially regulated by Epromoters. The pipeline requires a list of induced genes (RefSeq ID) and a ChIP-seq peak file (bed format) for a key transcription factor involved in the stress response.
For more details on the pipeline, please check our publication: Malfait et al. bioRxiv 2024.11.26.625372.